 Höpfler Lab
Höpfler Lab
 Genome Biology
Genome Biology
- Group page
- Research lines
- Group members
- Publications
Biosketch
2025 - Junior Group Leader, at the Centre for Genomic Regulation, Barcelona (Spain)
2021-2023 - Marie Skłodowska-Curie Postdoctoral Fellow, MRC Laboratory of Molecular Biology, Cambridge (UK)
2020-2021 - EMBO Postdoctoral Fellow, MRC Laboratory of Molecular Biology, Cambridge (UK)
2019-2024 - Postdoctoral Fellow, MRC Laboratory of Molecular Biology, Cambridge (UK); Advisor: Dr. Ramanujan Hegde
2019 - PhD in Biology, Ludwig-Maximilians-University Munich, and Max-Planck-Institute of Biochemistry, Martinsried (Germany); Advisors: Stefan Jentsch and Boris Pfander
2012-2013 - Research Associate, Medical University of Vienna (Austria); Advisor: Andrea Barta
2011 - Diploma in Molecular Biology,University of Vienna (Austria)
Research Summary
How do cells accurately tune protein production according to cellular needs? This fundamental question is not only fascinating but also crucial to human health: Aberrant protein levels are commonly linked to diseases such as developmental defects, cancer, or neurodegenerative diseases, where aggregation-prone proteins tend to accumulate in neurons.
In the “Dynamics of protein synthesis & RNA decay” lab we are interested in how cells tune protein production by adjusting the stability of messenger RNAs (mRNAs). Each cell in our body expresses around 12,000 different mRNAs at any given time. To control this complex transcriptome—and thus protein synthesis—cells adjust half-lives for individual mRNAs from minutes to several days. Traditionally, the selective degradation of mRNAs has been attributed to the recognition of nucleotide sequence elements by proteins or small RNAs that subsequently recruit decay factors. In our lab, we investigate a distinct, newly emerging paradigm of gene regulation termed “peptide-mediated mRNA decay” (PMD). In PMD, not the mRNA sequence but rather the nascent protein is recognized to trigger degradation of the encoding mRNA during its translation by the ribosome.
This process is exemplified by tubulin mRNAs, which are selectively degraded when cells sense excess free tubulins, the building blocks of the microtubule cytoskeleton. In this case, a recognition factor binds the nascent tubulin protein on the ribosome (see figure). Subsequently, an adapter protein and decay factors that execute mRNA degradation are recruited, as we and others recently demonstrated. Perturbed tubulin mRNA turnover results in severe phenotypes, such as cell division defects, impaired neurodevelopment, ciliopathies, and infertility. Beyond tubulins, emerging evidence points to critical PMD mechanisms targeting transcripts encoding aggregation-prone proteins when crucial chaperones are absent. Thus, PMD represents an unexplored paradigm of gene regulation that dynamically adjusts protein production to ensure cellular fitness. However, a mechanistic understanding of peptide-mediated mRNA decay is only beginning to emerge, and the scope of PMD substrates is completely unknown.
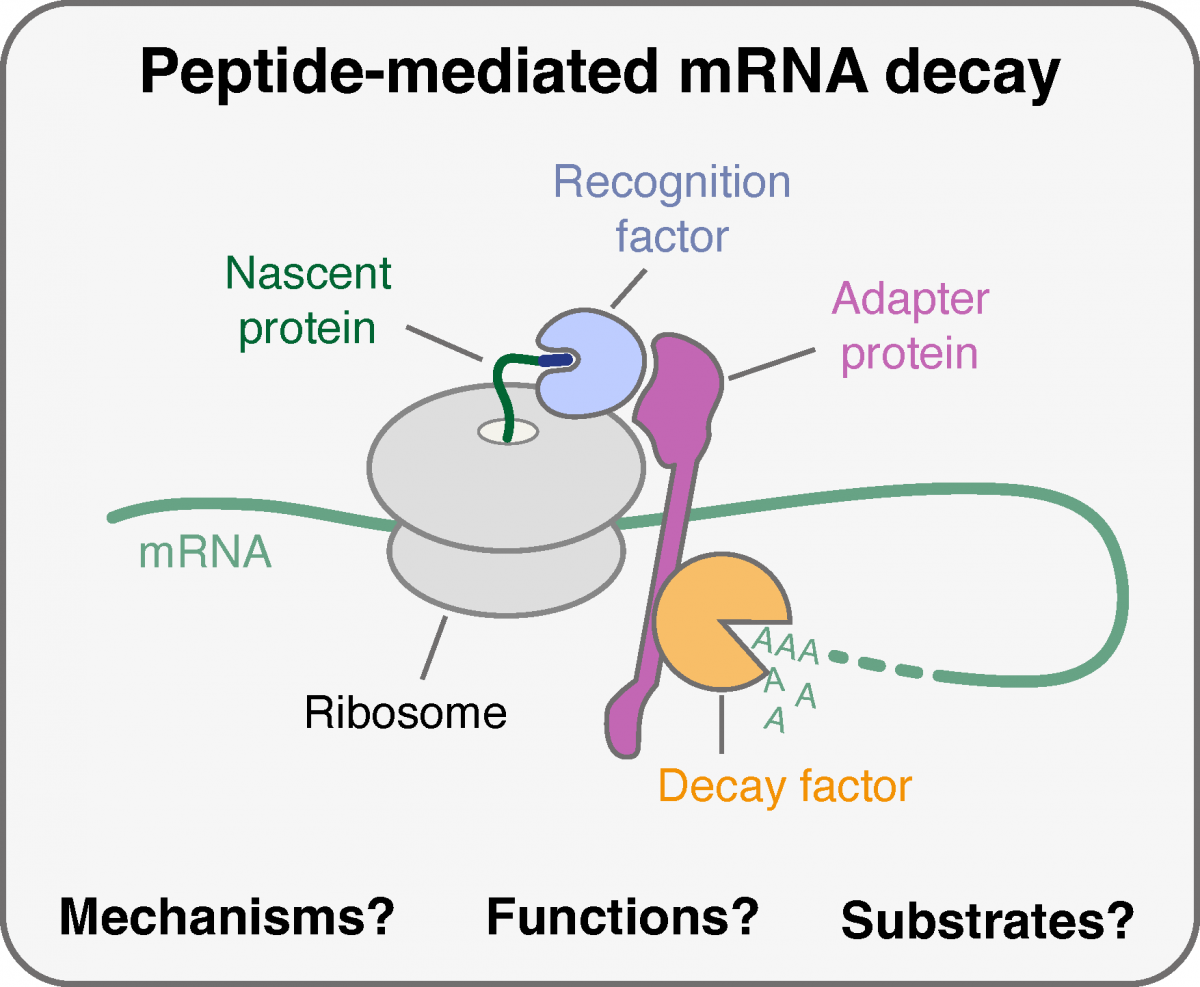
Using a highly interdisciplinary approach spanning biochemistry, structural biology, cell biology, genetics, and advanced sequencing methods, we aim to answer the following questions: What are the factors and molecular mechanisms driving peptide-mediated mRNA decay? How are these mechanisms controlled, and what are the consequences of impaired mRNA turnover for cells and organisms? What is the transcriptome-wide scope of regulation by peptide-mediated mRNA decay?
Answering these questions will be highly relevant to human biology and disease, as highlighted by the examples of tubulin and aggregation-prone proteins. Furthermore, insights into mRNA decay mechanisms will enable development of optimized mRNA-based therapeutics like mRNA vaccines.
Job Opportunities
We are always interested to hear from potential postdoctoral researchers, PhD students and technicians/research assistants that are interested in joining our lab. We are currently building an interdisciplinary team centred around biochemistry and cell biology, with some projects involving structural biology, next-generation sequencing or genetics approaches. Training in one or more of these disciplines and/or experience with RNA biology or microtubule biology will be beneficial. If you are excited about our research and are interested in joining the team, please reach out to Markus by email with a motivation letter and CV.
In our group, we aim to understand how cells use peptide-mediated mRNA decay (PMD) to tune protein production by directing mRNA degradation during its translation by the ribosome. Our research revolves around three key questions:
- What are the factors and molecular mechanisms driving peptide-mediated mRNA decay?
- How are these mechanisms controlled, and what are the consequences of impaired mRNA turnover for cells and organisms?
- What is the transcriptome-wide scope of regulation by peptide-mediated mRNA decay?
To address these questions, we employ two complementary strategies. First, we focus on unravelling the pivotal model pathway of tubulin autoregulation, where PMD is crucial to control the expression levels of alpha and beta tubulin subunits. Second, we aim to systematically identify novel substrates and mechanisms of PMD to deduce general principles of this novel type of post-transcriptional gene regulation.
Background
Tubulins are the building blocks of the microtubule (MT) cytoskeleton, which plays vital roles in cellular architecture, intracellular transport, and mitosis. Understanding MT regulation is highly relevant because aberrations of MTs are associated with a wide range of human pathologies, and they are a critical drug target in the therapy of cancer and other conditions. Importantly, the dynamic nature of MTs critically depends on the availability of free tubulin subunits. Around 40 years ago, it was recognized that when cells sense excess free tubulins, they trigger selective degradation of the encoding mRNAs to curb tubulin production. This process of “tubulin autoregulation” depends on the selective recognition of the nascent tubulin polypeptide on the ribosome.
Recently, the key factors and RNA decay steps for tubulin autoregulation have been identified (Fig. 1). TTC5 acts as a recognition factor for the nascent tubulin polypeptide on the ribosome (Lin et al., Science, 2020), and SCAPER in turn acts as an adapter molecule to recruit a general mRNA decay factor, the CCR4-NOT deadenylase complex (Höpfler et al., Mol Cell, 2023). Importantly, disease-linked SCAPER mutations abolish tubulin autoregulation and cause cell division defects. In patients, these SCAPER mutations cause a ciliopathy syndrome with neurodevelopmental defects, male infertility, and retinitis pigmentosa, allowing us to directly link the tubulin autoregulation pathway to human disease for the first time.
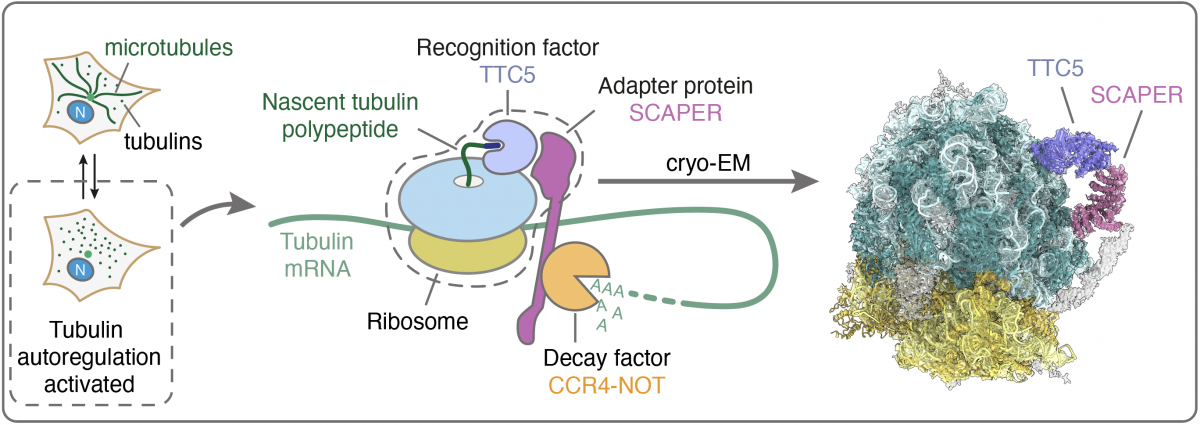
Figure 1. Schematic for the mechanism of peptide-mediated mRNA decay during tubulin autoregulation. Left: Tubulin autoregulation is activated when cells experience excess free tubulin subunits to curb tubulin production. Centre: Schematic of recognition, adapter, and decay factor assembly on tubulin-translating ribosomes. Right: Cryo-EM structure of the outlined complex. (Figure modified from Höpfler et al., Mol Cell, 2023)
Research lines
1.- Deciphering the coupling of mRNA translation and decay during tubulin PMD
We aim to decipher the molecular mechanisms and structural basis of how mRNA translation and decay are coupled for tubulin PMD. To achieve this, we use a range of biochemical methods such as cell-free translation systems and structural biology approaches to recapitulate tubulin PMD steps.
2.- Unravelling physiological triggers and regulation of tubulin PMD
We have recently identified the first regulatory step for the tubulin autoregulation pathway, where TTC5 is inhibited by a population of free tubulins (Batiuk, Höpfler, et al., Nat Commun, 2024). However, it remains unclear how other factors of the cascade are regulated, and what physiological signals trigger tubulin mRNA decay. We aim to identify and characterize regulatory factors and signalling pathways that control tubulin PMD and link them to endogenous triggers and disease phenotypes.
3.- Charting a transcriptome-wide map of PMD substrates and mechanisms
Beyond tubulin regulation, our findings raise the broader question of how PMD shapes human gene expression and the proteome. Which other mRNAs are selectively degraded through recognition of the nascent polypeptide during translation? Emerging evidence suggests that a wide range of mRNAs are subject to PMD-type regulation via the nascent polypeptide.
By innovating quantitative methods based on advanced sequencing technologies and reporter assays, we aim to overcome current limitations to study PMD regulation on a transcriptome-wide scale. Capitalizing on the methods and tools we employed for tubulin PMD, we will then dissect molecular mechanisms for the most relevant substrates. This will allow us to deduce general principles of PMD in gene regulation.


