 CRG INTERNATIONAL PhD PROGRAMME 2025 - CALL
CRG INTERNATIONAL PhD PROGRAMME 2025 - CALL

Apply for a PhD @ CRG
| Eligibility criteria | How to apply | Selection process | CRG PhD call timeline | Fellowships Available | Workshops | Contact
Apply now to boost your scientific career at the Centre for Genomic Regulation (CRG) in Barcelona. The CRG is a center of excellence with international teams representing a broad range of disciplines, with first-class core technologies to support the research projects, a wide range of seminars given by high-profile invited speakers, and courses on complementary and transferable skills integrated with the training programme.
Scroll down to learn more about 20 PhD fellowships available at the CRG
Deadline 12th January 2025
On the application form, you will be able to choose up to two research labs for which you wish to apply
Are you ready to prepare an outstanding application?
Join our online workshop to learn how to find the right lab for your PhD, prepare an outstanding application, and get expert tips to boost your chances of success!

GEBAUER'S LAB
RNA-Binding Protein | Metastasis | Persistence | Stress| Translation | Mechanisms
Our Research
The ‘Protein synthesis’ group headed by Fátima Gebauer studies how RNA-binding proteins contribute to translational reprogramming and stress resistance of cancer cells.
Who are we looking for?
We are looking for highly motivated researchers with background in RNA regulation. Strong interest in cancer biology, high-throughput technologies and bioinformatics is encouraged.

MALHOTRA'S LAB
Collagens | TANGO1 | Skin | Scleroderma | Aging
Our Research
The Intracellular Compartmentation Group, led by Vivek Malhotra, is focused on creating a synthetic dermal skin layer made from dermal fibroblasts, immune cells, and vascular cells. This engineered tissue will secrete extracellular matrix components, aiding in the investigation of skin aging and sclerosis mechanisms.
Who are we looking for?
Inquisitive researchers who are determined and passionate about science

FERRUZ'S LAB
Protein Design |Enzyme Engineering | Language Models |Multimodal Agents | New-to-Nature Design | Explainability
Our Research
Our lab aims to revolutionize controllable protein design by leveraging large language models, experimentally validating predictions, and refining models through feedback to enhance decision-making.
Who are we looking for?
PhD1: A computer scientist, mathematician, physician or computational biologist who will focus on the implementation of interpretability techniques (eg: influential functions, feature attribution methods) for protein language models.
PhD2: A chemist, biotechnologist, or biologist with wet-lab experience in structural biology, specifically protein expression, purification, thermodynamic and functional analaysis. The candidate will focus on the design and characterization of new-to-nature binders.

SURREY LAB
Physical Properties of the Cytosol | Active Networks | Self-organization | Crowding | Phase separation | Motor Protein | Microtubule
Our Research
The Surrey lab studies the design principles of biological self-organization, focusing on the cytoskeleton. Their interdisciplinary research merges synthetic biology, biophysics, biochemistry, cell biology and mathematical modelling.
Who are we looking for?
Master training in biophysics, biochemistry or bioengineering; a keen interest in complex dynamical systems; talent for protein biochemistry, biochemical in vitro reconstitutions, fluorescence/electron microscopy, and/or computational modelling.

BAUD LAB
Host-microbiome | Interactions | Metagenome-Assembled Genomes | Metabolome | Host Genetic Effects | Rodent Models
Our Research
The "Social and host-microbiome systems" group headed by Dr Amelie Baud aims to understand how the health of an individual can be affected not only by the individual's own genes but also by genes of social partners and the genes of the bacteria present in the individual’s microbiome.
Who are we looking for?
We are looking for a candidate keen to understand the biological mechanisms involved in host-microbiome interactions, with a mixed background in biology (ideally genetics or microbiology) and bioinformatics. Experience with R and/or Python as well as next generation sequencing data is necessary.

HÖPFLER'S LAB
Gene Regulation | Protein Synthesis | RNA Biology | Decay | Microtubules | In Vitro Reconstitution | Cell Division
Our Research
The “Dynamics of protein synthesis & RNA decay” group headed by Markus Höpfler investigates fundamental mechanisms of cellular protein and RNA homeostasis, focusing on disease-relevant RNA decay pathways.
Who are we looking for?
PhD1: Join us if you’re excited about revealing molecular mechanisms controlling RNA stability using biochemical reconstitution approaches. A strong background in structural/molecular biology, biochemistry or similar will be beneficial.
PhD2: Join us if you’re excited about investigating the crosstalk between cytoskeleton dynamics and mRNA regulation. A strong background in cell/molecular biology, gene regulation or genetics will be beneficial.

NOVOA'S LAB
RNA Biology | Ribosome Specialization | Cancer | Nanopore Sequencing | Disease Biomarkers | RNA Modifications | Antibiotic Resistance
Our Research
In our laboratory, we want to understand how gene expression is regulated with surgical precision, in a tissue-dependent, spatial and temporal dimension. Some of the key research lines of the lab include: i) dissect the role that RNA modifications play in sperm formation, maturation and intergenerational inheritance; ii) decipher how and why RNA modification dysregulation leads to human disease and antibiotic resistance ; and iii) bring direct RNA nanopore sequencing technologies towards the clinic, for early cancer diagnosis and stratification of patient samples.
Who are we looking for?
Enthusiastic candidate with hybrid wet and dry lab background that is interested in doing both molecular biology work and the corresponding data analysis, to try to understand why and how specialized ribosomses exist, what are their functions, and how we can exploit this knowledge to better understand antibiotic resistance mechanisms and design early cancer screening methodologies that rely on detection of oncogenic ribosomes.

LIN HSIU-CHUAN LAB
Single-Cell Sequencing & Multiomics | Cell Fate & State Engineering |Gene Regulation | Stem Cell-Derived Models | Biological Data Science | Perturbation Screening
Our Research
The "Cell Fate Decoding and Engineering" group led by Dr. Hsiu-Chuan Lin utilizes stem cell-derived models, perturbation screening, and single-cell multiomics to decode gene regulations underlying cell fate and state specification and use this knowledge to precisely engineer the identity of biomedically valuable human cells.
Who are we looking for?
We are seeking highly motivated researchers with a strong interest in gene regulation, cell fate transitions (e.g., in development, reprogramming, and diseases), cell engineering, and single-cell technology development. Candidates should have a background in life science, systems biology, bioengineering, or computational biology.
PhD1: Experience in stem cell models, genetic manipulation, and next-generation sequencing is required.
PhD2: Experience in biological data science and single-cell sequencing analysis is required. Experience in both wet and dry labs is a plus.

SEBÉ-PEDRÓS LAB
Comparative Transcriptomics | Genome Evolution | Population Genomics | Cis-Regulatory Evolution | Phylogenetics
Our Research
We study the origin and evolution of cell type programs and associated genome regulation mechanisms: transcription factors, 3D chromatin architecture, histone modifications, and more. To this end, we combine single-cell genomics, chromatin profiling, and phylogenetic comparative methods in diverse animals and unicellular eukaryotes.
Who are we looking for?
We are looking for candidates broadly interested in evolutionary biology, gene regulation, and comparative genomics; and with a background in genetics, functional genomics and/or computational biology.

BEEKMAN LAB
LLymphoma | Genome Regulation and Organization | Translocations | Single-cell Multiomics | DNA Methylation
Our Research
Our lab focuses on the understanding of normal B-cell differentiation and lymphoma formation from the perspective of 3D chromatin organization, DNA methylation and single-cell heterogeneity in healthy and pre-malignant lymphocyte cell populations.
Who are we looking for?
We are looking for two enthusiastic candidates, with respective hands on experience in wetlab or bioinformatics for at least 1 year, to pursue a wetlab-oriented project regarding gene regulation in lymphoma formation, and a single-cell multiomics bioinformatic project to identify new B-cell subtypes and their connection to lymphoma formation.

VELTEN'S LAB
Single cell genomics | Deep learning / Bioinformatics | Stem Cells | Gene Regulation | Ageing
Our Research
The Velten lab uses single-cell and synthetic genomics, high-throughput genetic screens and artificial intelligence to study the genomic regulation of differentiation programs in hematopoietic and leukemic stem cells. Currently, important topics in the lab are clonal dynamics in blood during ageing, as well as the genomic encoding of differentiation programs.
Who are we looking for?
We are looking for a motivated PhD student with a strong interest in gene regulation and stem cell biology. Both bioinformatic and wetlab profiles are welcome to apply, but ideally, you should have experience in both disciplines.

COSMA LAB
Stem Cells | Chromatin | Super Resolution Imaging | Genome Organisation
Our Research
We are investigating the functional role of 3D genome organization using super resolution microscopy, genomic and modeling approaches. Our research is focused on elucidating how nuclear architecture and chromatin looping regulate cellular function in stem cells, somatic cells and cancer.
Who are we looking for?
We are looking for highly motivated individuals with a degree in biology, biotechnology, biophysics or related disciplines with expertise in imaging, cellular, and molecular biology.

LATORRE'S LAB
Aging | Epigenetics | Cellular dynamics | Clonal Haematopoiesis | Mathematical Modelling
Our Research
Our interdisciplinary group uses experimental and mathematical modeling to explore aging patterns and their link to age-related diseases. Guided by both fundamental and clinical questions, we aim to understand: What cellular processes drive aging? Can we develop interpretable measures of aging to better understand its role in disease? And how can this knowledge aid in early disease detection and extending healthspan?
Who are we looking for?
We are seeking for a candidate with existing expertise in mathematical and computational techniques, or a strong willingness to learn these skills, to work on developing quantitative models of stem cell dynamics related to the aging process.

VERNÓS LAB
Cytoskeleton | Microtubules | Cell Division | Cancer | Aneuploidy
Our Research
The Vernos Group aims at understanding how the bipolar spindle segregates faithfully the chromosomes during mitosis and meiosis and the mechanisms that may be at stake in pathologies such as infertility and cancer.
Who are we looking for?
We are looking for a highly motivated researcher with a background in cell and cancer biology and solid skills in quantitative cell biology and advanced optical microscopy methods.

BÖKE LAB
Oocyte | Protein Homeotasis |Fertility
Our Research
The main focus of our lab is to reveal the mechanisms oocytes, female germ cells that become eggs, employ to establish homeostasis during their long lifespan, and why these mechanisms eventually fail with advanced age. If you are interested in understanding how life begins, apply!
Who are we looking for?
The candidate is enthusiastic about understanding the mysteries of life! Experience in cell and developmental biology is a big plus.

STROUSTRUP LAB
Aging |Biomarker | Quantitative Imaging | Statistical modelling | Machine Learning | Molecular Genetics
Our Research
Our group studies the molecular origins of aging, working to understand how functional declines at the molecular level can lead to emergent aging phenotypes at the organismal-level. We ask questions like: why do some individuals live longer than other? Why do point mutations in the insulin receptor double lifespan? We are an interdisciplinary group, combining experimental methods taken from functional genetics, systems biology, and CRISPR genome editing with a variety of statistical and phenomenological modelling techniques.
Who are we looking for?
We are looking for candidates interested in experimental research on aging, with a focus on biomarkers, quantitative measurement, and interdisciplinary projects working closely with bioinformaticians and engineers. We are particularly interested in students with training in experimental life sciences or engineering.

WEGHORN LAB
Computational Biology | Evolution | Genomics |Mutation Rates | Darwinian Selection
Our Research
The Weghorn group works on computational and probabilistic approaches to modeling mutation rates and selection pressures acting on evolving genomes. We are particularly interested in cancer and human evolution.
Who are we looking for?
We are looking for a candidate with a quantitative background interested in computationally studying large sequencing datasets using evolutionary models and statistical analysis.

IRIMIA LAB
Viral infection | Comparative transcriptomics | Translatomics | Chikungunya virus
Our Research
We use comparative functional genomics to understand how organisms evolve and why we are susceptible to diseases. This project is in co-supervision with Juana Diez at UPF, as part of the Evolutionary Medical Genomics Program, blending molecular virology and evolutionary biology.
Who are we looking for?
A passionate researcher with a bioinformatics background excited about applying evolutionary principles to understand human diseases.
Eligibility criteria
Candidates should fulfill the following eligibility criteria at the time of the call deadline. If it becomes clear before, during, or after the evaluation phase that one or more of the eligibility criteria have not been fulfilled, the proposal will be declared ineligible and withdrawn from further examination.
- The call is open to candidates of any nationality.
- Candidates must have obtained a University Bachelor's Degree and a Master's Degree in biomedical sciences within the European Higher Education System (minimum 300 ECTS) or an equivalent University Degree that allows them to start a PhD thesis in Spain. Candidates who expect to be awarded with such a degree by September 2025 are eligible to apply.
- Candidates must have an excellent academic record, previous research experience, and a strong commitment to scientific research.
- Candidates must have a high working knowledge of English.
- Candidates invited to previous CRG PhD call interviews are not eligible.
- Candidates performing their Masters at the CRG are not eligible.
- Candidates may not have worked at the CRG for more than 3 months before the call deadline.
How to apply
- Applications must be submitted online - no applications by email will be considered.
- Candidates must register to use the online application system.
- The online application form requests all of the necessary information for the initial stages of the selection process (General information, education, academic transcripts, scholarships, prizes and awards, research experience, scientific interests and reference letters).
- If the academic transcripts are not in Catalan, Spanish or English applicants should also attach a translation in one of the above-mentioned languages.
- Two reference letters will be automatically requested from the referees proposed by the candidate through the online system.
- Applicants should select a maximum of 2 CRG labs in which they would like to start a PhD.
- Candidates must ensure that all information and documents are included before the deadline, including the reference letters. Incomplete proposals will not be considered.
Once the application is submitted, an acknowledgment of receipt will be automatically sent by e-mail to the applicant.
For any additional information, please check the FAQs.
Selection process
The selection of the fellows will be based on the candidates' academic qualifications and research excellence.
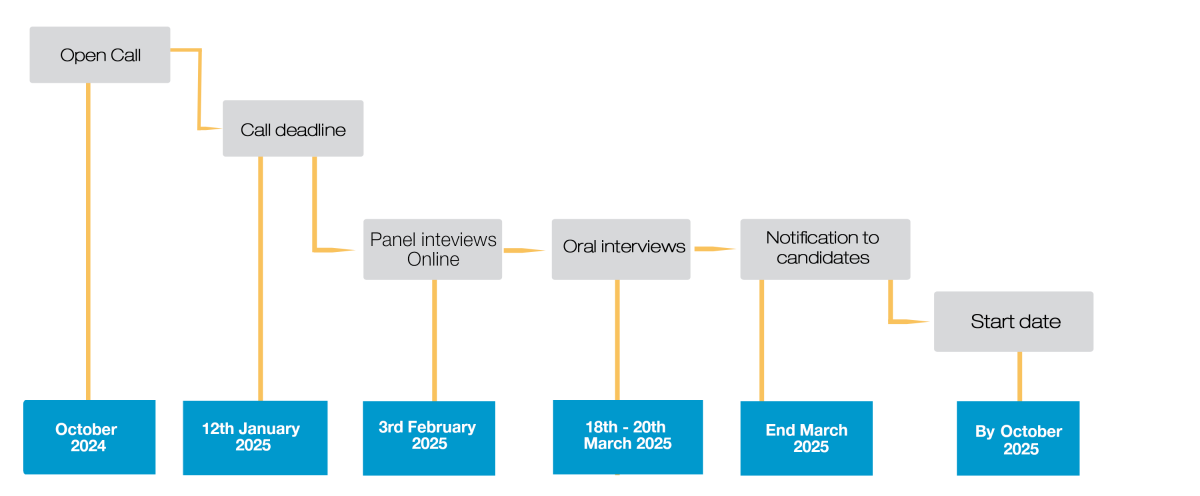
- Pre-selection: The pre-selection will be based on the candidate's application, reference letters, research interests, motivation, and fit to the selected labs. If the candidate is pre-selected, she/he will be invited to an online panel interview via Zoom on the 3rd of February 2025. If successfully passes the panel interview, the candidate will be invited to Barcelona for the presential interviews.
- Presential interviews: The interviews will take place from the 18th to the 20th of March 2025 at the CRG in Barcelona. Accommodation and flight tickets will be provided. Please be aware that during all those days, interviews and presentations will take place. Full details regarding the interview process will be sent to invited candidates.
- Notification to candidates: Offers will be made to the successful candidates shortly after the interview. Successful candidates would be expected to start the PhD latest by October 2025.
CRG PhD Call Timeline
Fellowships available
Workshops
We are organizing two online events to discuss what to take into consideration when looking for a PhD position and how to prepare an outstanding application. These workshops will be delivered by the Training & Academic Office (TAO).
To participate in the event registration is needed.
ONLINE WORKSHOP - Envision Your Research Future – How to choose a lab for your PhD & CRG opportunities
- Thursday 21st November 2024 - 10am CET - REGISTRATION HERE
- Friday 13th December 2024 - 3pm CET - REGISTRATION HERE
Contact
For any additional information, please contact the CRG Training & Academic Office at: training@crg.eu
