 Genome Research (2005): Comparison of Splice Sites in Mammals and Chicken
Genome Research (2005): Comparison of Splice Sites in Mammals and Chicken
SUPPLEMENTARY MATERIALS FOR
Comparison of Splice Sites in Mammals and Chicken
J. F. Abril, R. castelo, and R. Guigó *.
Genome Research, 15(1):111-119, January 3, 2005.
[PubMed] [Abstract] [Full Text] [Datasets]
[Published online before print in Dec 2004]
* To whom correspondence should be addressed.
Contact Author. Ph: +34 93 225 7567.
Contents
Summary
We have carried out an initial analysis of the dynamics of the recent evolution of the splice sites sequences on a large collection of human, rodent (mouse and rat), and chicken introns. Our results indicate that the sequences of splice sites are largely homogeneous within tetrapoda. We have also found that orthologous splice signals between human and rodents and within rodents are more conserved than unrelated splice sites, but the additional conservation can be explained mostly by background intron conservation. In contrast, additional conservation over background is detectable in orthologous mammalian and chicken splice sites. Our results also indicate that the U2 and U12 intron classes seem to have evolved independently since the split of mammals and birds; we have not been able to find a convincing case of interconversion between these two classes in our collections of orthologous introns. Similarly, we have not found a single case of switching between AT-AC and GT-AG subtypes within U12 introns, suggesting that this event has been a rare occurrence in recent evolutionary times. Switching between GT-AG and the non-canonical GC-AG U2 subtypes, on the contrary, does not appear to be unusual; in particular, T to C mutations appear to be relatively well tolerated in GT-AG introns with very strong donor sites.
UCSC Initial RefSeq Datasets
RefSeq Identifiers from Filtered Sets
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Hsap | UCSC_200307 | 21744 | 20894 | 18117 | 15159 | 10757 | 7799 | 17939 | 15066 | 10316 | 7443 | 21091 |
| Mmus | UCSC_200310mm | 17988 | 16126 | 14432 | 13677 | 9765 | 9010 | 14175 | 13461 | 9078 | 8364 | 16192 |
| Rnor | UCSC_200306rn | 4798 | 4134 | 3454 | 3347 | 2201 | 2094 | 3368 | 3275 | 1947 | 1854 | 4536 |
| Ggal | UCSC_200402 | 1496 | 1085 | - | - | - | - | - | - | - | - | 1367 |
| Hsap | UCSC_20030410 | 19174 | 18337 | 18145 | 18067 | 10486 | 10408 | 18014 | 17901 | 9988 | 9875 | 18226 |
| Mmus | UCSC_200302mm | 13406 | 11161 | 10503 | 10404 | 7397 | 7298 | 10371 | 10255 | 6908 | 6792 | 12511 |
| Rnor | UCSC_200301rn | 4219 | 3372 | 3070 | 3049 | 2102 | 2081 | 3017 | 2991 | 1893 | 1867 | 4002 |
1.- Total RefSeqs
2.- (1) without Stop codons in frame when translating from genomic
3.- (2) + (identity(aa)>95% + gap(aa)<6) or (identity(RNA)>95% + gap(RNA)<16)
4.- (2) + (identity(aa)>95% + gap(aa)<6)
5.- (2) + (identity(RNA)>95% + gap(RNA)<16)
6.- (2) + (identity(aa)>95% + gap(aa)<6) and (identity(RNA)>95% + gap(RNA)<16)
7.- (2) + (mismatch(aa)<4 + gap(aa)<6) or (mismatch(RNA)<10 + gap(RNA)<16)
8.- (2) + (mismatch(aa)<4 + gap(aa)<6)
9.- (2) + (mismatch(RNA)<10 + gap(RNA)<16)
10.- (2) + (mismatch(aa)<4 + gap(aa)<6) and (mismatch(RNA)<10 + gap(RNA)<16)
11.- Unique ID
Sequence Files for All RefSeq Genes: Exons, Introns, CDS and Splice Sites.
| Based on | All Exons | All Introns | All CDSs | Splice Sites | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| refgenes.txt | SEQ(fasta) | SEQ(content) | SEQ(fasta) | SEQ(content) | SEQ(fasta) | SEQ(content) | EXONIC | INTRONIC | |||
| Hsap UCSC200307 | 19M | 3.7M | 362M | 4.9M | 11M | 4.8M | 19M | 17M | |||
| Mmus UCSC200310 | 15M | 2.8M | 211M | 3.6M | 8.5M | 3.7M | 15M | 14M | |||
| Rnor UCSC200306 | 4.0M | 878K | 70M | 1.1M | 2.6M | 1.1M | 4.7M | 4.4M | |||
| Ggal UCSC200402 | 1.2M | 260K | 13M | 325K | 772K | 328K | 1.4M | 1.3M | |||
| Hsap UCSC200304 | 16M | 3.3M | 304M | 4.3M | 9.3M | 4.3M | 17M | 16M | |||
| Mmus UCSC200302 | 10M | 2.1M | 141M | 2.7M | 6.4M | 2.7M | 12M | 11M | |||
| Rnor UCSC200301 | 3.4M | 751K | 55M | 1M | 2.3M | 962K | 4.0M | 3.7M | |||
This table shows the file sizes of the gzipped files in each category.
Click on file size numbers to retrieve the corresponding file.
RefSeq U2/U12 Intron Major Classes
Summary of U2/U12 Intron Major Classes on RefSeq Filtered Set 1 (Total RefSeqs)
| U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | ||||||||
| Hsap UCSC200307 | 189656 | 1529 | 34 | 2248 | 128 | 2 | 9 | 8 | 12430 | 109 | 13 | 134 | 355 | 1 | 139 | 19 | 206814 | ||||||
| Mmus UCSC200310 | 125587 | 1015 | 21 | 2407 | 88 | 0 | 7 | 9 | 9557 | 66 | 10 | 130 | 254 | 1 | 91 | 15 | 139258 | ||||||
| Rnor UCSC200306 | 38601 | 289 | 14 | 1236 | 20 | 0 | 1 | 1 | 3038 | 19 | 4 | 77 | 69 | 0 | 20 | 4 | 43393 | ||||||
| Ggal UCSC200402 | 11073 | 77 | 5 | 736 | 7 | 0 | 1 | 0 | 676 | 6 | 0 | 27 | 17 | 0 | 5 | 2 | 12632 | ||||||
| Hsap UCSC200304 | 162740 | 1254 | 28 | 2273 | 115 | 0 | 9 | 6 | 10846 | 91 | 13 | 126 | 302 | 1 | 108 | 19 | 177931 | ||||||
| Mmus UCSC200302 | 92487 | 721 | 16 | 3740 | 69 | 0 | 6 | 9 | 7027 | 46 | 5 | 192 | 196 | 1 | 67 | 9 | 104591 | ||||||
| Rnor UCSC200301 | 32378 | 253 | 13 | 1589 | 18 | 0 | 1 | 2 | 2604 | 17 | 3 | 82 | 60 | 0 | 20 | 3 | 37043 | ||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
| Search parameters:
|
donor_pattern |
= | /^ATCCT[CT]/ |
|---|---|---|---|
acceptor_max_mismatch_number |
= | 1 |
|
acceptor_pattern |
= | /TCCTT[AG]AC/ |
| U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | ||||||||
| Hsap UCSC200307 | 190632 | 1536 | 34 | 2249 | 128 | 2 | 9 | 8 | 11454 | 102 | 13 | 133 | 355 | 1 | 139 | 19 | 206814 | ||||||
| Mmus UCSC200310 | 126409 | 1021 | 21 | 2408 | 89 | 0 | 7 | 9 | 8735 | 60 | 10 | 129 | 253 | 1 | 91 | 15 | 139258 | ||||||
| Rnor UCSC200306 | 38848 | 289 | 14 | 1238 | 20 | 0 | 1 | 1 | 2791 | 19 | 4 | 75 | 69 | 0 | 20 | 4 | 43393 | ||||||
| Ggal UCSC200402 | 11150 | 78 | 5 | 736 | 7 | 0 | 1 | 0 | 599 | 5 | 0 | 27 | 17 | 0 | 5 | 2 | 12632 | ||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
| Search parameters:
|
donor_pattern |
= | /^ATCCT[CT]/ |
|---|---|---|---|
acceptor_max_mismatch_number |
= | 2 |
|
acceptor_pattern |
= | /TCCTT[AG]AC/ |
| U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | ||||||||
| Hsap UCSC200307 | 108118 | 973 | 21 | 1571 | 34 | 0 | 3 | 3 | 93968 | 665 | 26 | 811 | 449 | 3 | 145 | 24 | 206814 | ||||||
| Mmus UCSC200310 | 69628 | 567 | 13 | 1647 | 22 | 0 | 1 | 2 | 65516 | 514 | 18 | 890 | 320 | 1 | 97 | 22 | 139258 | ||||||
| Rnor UCSC200306 | 20943 | 168 | 9 | 855 | 4 | 0 | 0 | 0 | 20696 | 140 | 9 | 458 | 85 | 0 | 21 | 5 | 43393 | ||||||
| Ggal UCSC200402 | 6444 | 49 | 4 | 600 | 0 | 0 | 0 | 0 | 5305 | 34 | 1 | 163 | 24 | 0 | 6 | 2 | 12632 | ||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
| Search parameters:
|
donor_pattern |
= | /^ATCCT[CT]/ |
|---|---|---|---|
acceptor_max_mismatch_number |
= | 2 |
|
acceptor_pattern |
= | /TCCTT[AG]AC/ |
|
| Extra constraints:
|
branchpoint_distance_from_acceptor |
= | [ -20 .. -5 ] |
branchpoint_sequence_matches_to |
= | [ /..A.$/ || /.A..$/ ] |
| U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | ||||||||
| Hsap UCSC200307 | 182013 | 1471 | 31 | 2127 | 51 | 0 | 3 | 4 | 20073 | 167 | 16 | 255 | 432 | 3 | 145 | 23 | 206814 | ||||||
| Mmus UCSC200310 | 120700 | 968 | 20 | 2316 | 32 | 0 | 1 | 2 | 14444 | 113 | 11 | 221 | 310 | 1 | 97 | 22 | 139258 | ||||||
| Rnor UCSC200306 | 37208 | 275 | 14 | 1204 | 8 | 0 | 0 | 0 | 4431 | 33 | 4 | 109 | 81 | 0 | 21 | 5 | 43393 | ||||||
| Ggal UCSC200402 | 10698 | 76 | 5 | 733 | 2 | 0 | 0 | 0 | 1051 | 7 | 0 | 30 | 22 | 0 | 6 | 2 | 12632 | ||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
U2/U12 Pictograms
| CLASS | DONOR SITES | ACCEPTOR SITES |
|---|---|---|
| GT-AG | 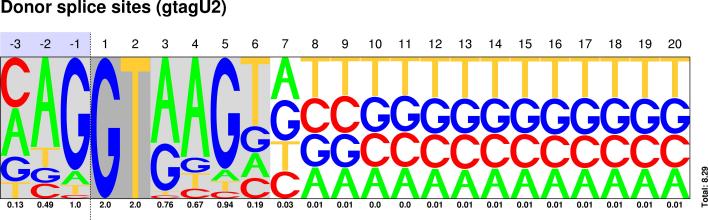 Sequences : PWM : JPG / PNG / PS |
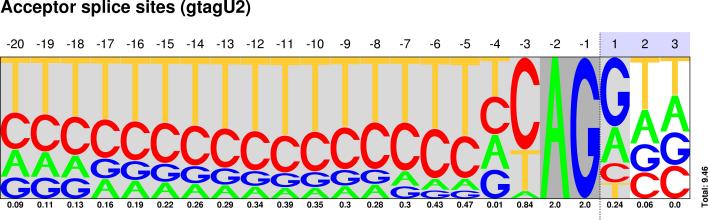 Sequences : PWM : JPG / PNG / PS |
| GC-AG |  Sequences : PWM : JPG / PNG / PS |
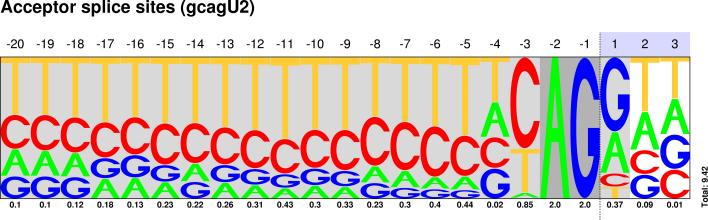 Sequences : PWM : JPG / PNG / PS |
| U12 | 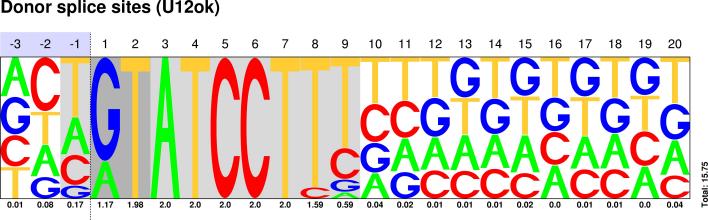 Sequences : PWM : JPG / PNG / PS |
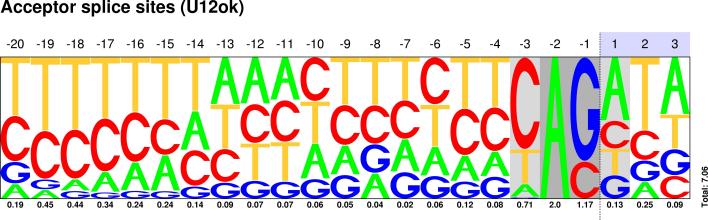 Sequences : PWM : JPG / PNG / PS |
| BRANCH POINT |
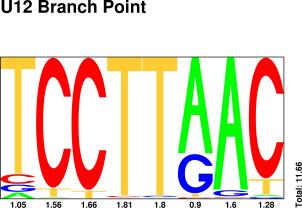 Sequences : PWM : JPG / PNG / PS |
|
U2/U12 Splice Sites Datasets
Summary of U2 Intron Major Classes on RefSeq Orthologous Set (Paper Table 3)
| U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | ||||||||
| Hsap UCSC200307 | 31425 | 218 | 3 | 29 | 27 | 0 | 0 | 2 | 2055 | 12 | 0 | 7 | 4 | 0 | 1 | 0 | 33783 | ||||||
| Mmus UCSC200310 | 28168 | 207 | 2 | 70 | 23 | 0 | 0 | 0 | 2231 | 14 | 1 | 9 | 2 | 0 | 0 | 0 | 30727 | ||||||
| Rnor UCSC200306 | 10019 | 64 | 4 | 23 | 5 | 0 | 0 | 1 | 835 | 9 | 0 | 5 | 0 | 0 | 0 | 0 | 10965 | ||||||
| Hsap UCSC200304 | 31626 | 220 | 3 | 28 | 27 | 0 | 0 | 2 | 2068 | 12 | 0 | 6 | 2 | 0 | 0 | 0 | 33994 | ||||||
| Mmus UCSC200302 | 28810 | 212 | 2 | 41 | 24 | 0 | 0 | 0 | 2270 | 14 | 0 | 7 | 3 | 0 | 0 | 0 | 31383 | ||||||
| Rnor UCSC200301 | 10209 | 65 | 4 | 7 | 5 | 0 | 0 | 1 | 841 | 9 | 0 | 4 | 0 | 0 | 0 | 0 | 11145 | ||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
Summary of U12 Intron Major Classes on RefSeq Orthologous Set
| U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | |||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | GTAG | GCAG | ATAC | XXXX | ||||||||
| Hsap UCSC200307 | 2 | 0 | 0 | 0 | 9 | 0 | 0 | 1 | 7 | 0 | 1 | 1 | 65 | 0 | 31 | 0 | 117 | ||||||
| Mmus UCSC200310 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 7 | 0 | 2 | 1 | 71 | 0 | 27 | 1 | 114 | ||||||
| Rnor UCSC200306 | 1 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 26 | 0 | 9 | 0 | 39 | ||||||
| Hsap UCSC200304 | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 1 | 7 | 0 | 1 | 1 | 67 | 0 | 31 | 0 | 118 | ||||||
| Mmus UCSC200302 | 1 | 0 | 0 | 0 | 2 | 0 | 2 | 0 | 6 | 0 | 2 | 1 | 73 | 0 | 28 | 1 | 116 | ||||||
| Rnor UCSC200301 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 27 | 0 | 9 | 0 | 39 | ||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
Orthologous U2/U12 Splice Sites
Chicken Orthologous for Human/Mouse/Rat U12 Splice Sites
| x Gg200402 | U2 Both Sites | U12 Donor Site | U12 Acceptor Site | U12 Both Sites | TOTAL | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Exon- erate |
Genic CDS |
GT AG |
GC AG |
AT AC |
XX XX |
GT AG |
GC AG |
AT AC |
XX XX |
GT AG |
GC AG |
AT AC |
XX XX |
GT AG |
GC AG |
AT AC |
XX XX |
||||||||||
| Hs200307/Mm200310/Rn200306 | TBL | FA | GFF | 1 | 2 | 0 | 27 | 9 | 0 | 0 | 0 | 4 | 2 | 0 | 5 | 29 | 0 | 8 | 2 | 89 | |||||||
| Hs200304/Mm200302/Rn200301 | TBL | FA | GFF | 1 | 2 | 0 | 28 | 9 | 0 | 0 | 0 | 5 | 2 | 0 | 6 | 30 | 0 | 8 | 3 | 94 | |||||||
Click on numbers of the column TOTAL to retrieve the table with the splice sites info.
| Exonerate parameters:
|
--query NNN.u12.exoncdspairs.fa |
(where NNN was hsap.gp200307, mmus.gp200310, rnor.gp200306 hsap.gp200304, mmus.gp200302, or rnor.gp200301) |
|---|---|---|
--target chromfa/chrNNN.fa |
(where NNN is a chicken chromosome number from this table) | |
--softmasktarget |
||
--model coding2genome |
||
--proteinsubmat blosum62 |
Alignments Summaries for the Orthologous U12 Splice Sites Comparison
| Orthologous Intron IDs Hsap/Mmus/Rnor Hsap/Mmus Hsap/Rnor Mmus/Rnor Hsap/Ggal Mmus/Ggal Rnor/Ggal Orthologous Introns IDs Orthologous Splice Sites Alignments
|
|
||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
Orthologous Human/Mouse/Rat U12 Introns Alignments against Chicken.
Comparative Pictograms
Sequence Files for Comparative Analysis of Splice Sites.
| Site Sequences | |||||||
|---|---|---|---|---|---|---|---|
| Species | Data Sets | Donors (-3/GT/+4) |
Acceptors (-18/AG/+3) |
||||
| H.sapiens | seq.gz | dat.gz | dat.gz | ||||
| M.musculus | seq.gz | dat.gz | dat.gz | ||||
| R.norvegicus | seq.gz | dat.gz | dat.gz | ||||
| G.gallus | seq.gz | dat.gz | dat.gz | ||||
| D.rerio | fasta.gz | dat.gz | dat.gz | ||||
| D.melanogaster | fasta.gz | dat.gz | dat.gz | ||||
Comparative Pictograms for Donor and Acceptor Splice Sites.
| Species | Donor Sites | Acceptor Sites |
|---|---|---|
| M.musculus R.norvegicus |
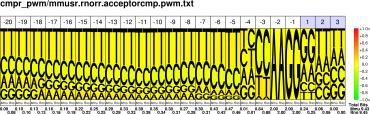 PWM : JPG / PNG / PS |
|
| H.sapiens M.musculus |
 PWM : JPG / PNG / PS |
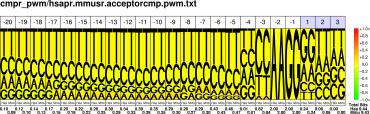 PWM : JPG / PNG / PS |
| H.sapiens R.norvegicus |
 PWM : JPG / PNG / PS |
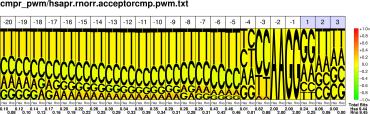 PWM : JPG / PNG / PS |
| H.sapiens G.gallus |
 PWM : JPG / PNG / PS |
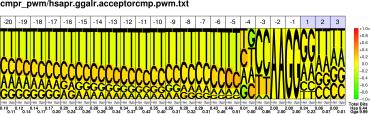 PWM : JPG / PNG / PS |
| H.sapiens D.rerio |
 PWM : JPG / PNG / PS |
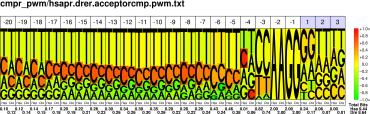 PWM : JPG / PNG / PS |
| H.sapiens D.melanogaster |
 PWM : JPG / PNG / PS |
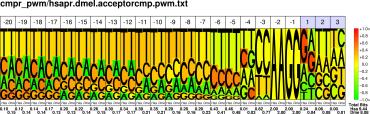 PWM : JPG / PNG / PS |
Sequence Conservation
To perform the following analyses we started from a set of human, mouse, rat and chicken, reliable 1:1:1:1 orthologs, kinly provided by Peer Bork and Ivica Letunic as part of the International Chicken Genome Sequencing Consortium (ICGSC) collaborations. From that set we produced the file linked below, containing the 1:1:1:1 orthologous introns for which the donor and acceptor sites used in the conservation analysis were retrieved.
orthointrons_sites.hmrg.tbl contains 6524 orthologous introns.
Sequence Files for All UCSC Ensembl Genes: Exons, Introns, CDS and Splice Sites.
| Based on | All Exons | All Introns | All CDSs | Splice Sites | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| ensgenes.txt | SEQ(fasta) | SEQ(content) | SEQ(fasta) | SEQ(content) | SEQ(fasta) | SEQ(content) | EXONIC | INTRONIC | |||
| Hsap UCSC200307 | 22M | 5.0M | 507M | 6.2M | 24M | 6.2M | 22M | 20M | |||
| Mmus UCSC200310 | 20M | 4.2M | 293M | 5.1M | 21M | 5.3M | 19M | 18M | |||
| Rnor UCSC200306 | 13M | 3.7M | 234M | 4.4M | 14M | 4.1M | 17M | 15M | |||
| Ggal UCSC200402 | 13M | 4.1M | 180M | 4.7M | 14M | 4.6M | 18M | 16M | |||
This table shows the file sizes of the gzipped files in each category.
Click on file size numbers to retrieve the corresponding file.
Sequence Datasets for Donor and Acceptor Orthologous Splice Sites.
| Donor Sites | Acceptor Sites | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Species | Orthologous Pairs |
Identity Summary |
Random Pairs |
Identity Summary |
Orthologous Pairs |
Identity Summary |
Random Pairs |
Identity Summary |
|||||||
| M.musculus | / | R.norvegicus | TBL | SET | TBL | SET | TBL | SET | TBL | SET | |||||
| H.sapiens | / | M.musculus | TBL | SET | TBL | SET | TBL | SET | TBL | SET | |||||
| H.sapiens | / | R.norvegicus | TBL | SET | TBL | SET | TBL | SET | TBL | SET | |||||
| H.sapiens | / | G.gallus | TBL | SET | TBL | SET | TBL | SET | TBL | SET | |||||
| G.gallus | / | M.musculus | TBL | SET | TBL | SET | TBL | SET | TBL | SET | |||||
| G.gallus | / | R.norvegicus | TBL | SET | TBL | SET | TBL | SET | TBL | SET | |||||
| Human/mouse/rat/chicken orthologous introns file: TBL | |||||||||||||||

