 NOTICIAS
NOTICIAS
This one goes up to 11: researchers crack code for genetic ‘control dials’
PRESS RELEASE
This one goes up to 11: researchers crack code for genetic ‘control dials’
Scientists at the Centre for Genomic Regulation (CRG) in Barcelona, Spain, have developed a new technique to crack the underlying DNA code for the ‘control dials’ that determine levels of gene activity in bacteria. The discovery has important implications for biotechnology, because genetically engineered bacteria and other organisms are used to produce useful molecules such as new materials and drugs.
When a gene is switched on, various stretches of DNA nearby act as ‘control dials’, influencing the level of activity and the amount of gene product that is made. Using Mycoplasma pneumoniae bacteria as a model, CRG director Professor Luis Serrano and his team developed a rapid way of scanning through many thousands of randomly-generated DNA sequences in search of those that could efficiently activate a ‘reporter’ gene.
The researchers used the new technique, known as ELM-seq, to find DNA sequences that strongly increase the levels of transcription – the process by which a gene is ‘read’ to produce an intermediate message known as RNA. They also searched for sequences that enhance the efficiency of translation, when RNA messages are interpreted to build products such as protein molecules. The patented ELM-seq method relies on indirectly measuring the activity of a gene encoding a protein that adds a specific chemical ‘tag’ on to DNA. If the gene is more active it will leave more tags on the DNA. These tags are detected and measured using a sensitive DNA analysis technique known as massive, parallel sequencing, providing a quantitative readout of the gene’s activity levels.
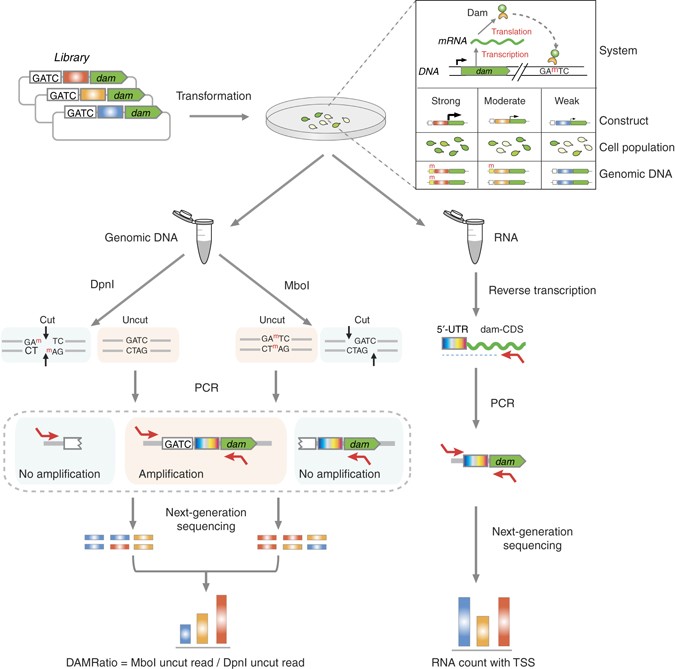 Publishing their findings in the open-access journal Nature Communications, the researchers discovered DNA sequences for ‘control dials’ that consistently produce very high levels of gene activity. They also revealed previously unknown information about the kinds of DNA sequences that work best. Intriguingly, the team discovered that the very first ‘letter’ (base) of the RNA message is very important for efficient gene transcription. They also found that the three-dimensional structure of the RNA message plays a key role in determining how well the RNA will be translated to make proteins, rather than specific sequences that were previously thought to be essential for efficient translation.
Publishing their findings in the open-access journal Nature Communications, the researchers discovered DNA sequences for ‘control dials’ that consistently produce very high levels of gene activity. They also revealed previously unknown information about the kinds of DNA sequences that work best. Intriguingly, the team discovered that the very first ‘letter’ (base) of the RNA message is very important for efficient gene transcription. They also found that the three-dimensional structure of the RNA message plays a key role in determining how well the RNA will be translated to make proteins, rather than specific sequences that were previously thought to be essential for efficient translation.
“Previous techniques that have been used to investigate DNA control sequences usually rely on sorting cells one by one and measuring gene activity in each of them,” says Dr Eva Yus, lead author of the paper. “However, our new approach uses cutting-edge DNA sequencing technology to precisely measure the effects of thousands of sequences on gene activity at the same time.”
Mycoplasma pneumoniae has a very small genome, which makes it relatively easy to study. But while this study has proved that the technique works in a simple organism, it could also be applied to other bacterial species, yeast or even human cells to find useful information about how genes are controlled and how they can be manipulated.
“If we want to use bacteria or other cells for biotechnology applications, we need to engineer them to make the maximum amount of product. But it’s very difficult if we don’t have information about the optimum sequences for controlling genes,” says Dr Jae-Seong Yang, co-author of the study.
Because Mycoplasma pneumoniae normally infects the lungs, the team now plans to use their new knowledge to develop therapies based on genetically engineered bacteria for treating lung diseases. Their targets include lung infections, cancer, and even approaches for regenerating damaged tissue.
“We now have a big range of control sequences we can use to adjust the levels of the proteins we want to produce in the lungs – like a toolkit we can use to control gene activity,” Professor Serrano explains. “In addition, this technique is a very cheap and fast way of finding the sequences governing transcription and translation, and could be used for any biotechnology application or organism.”
Reference:
Eva Yus, Jae-Seong Yang, Adrià Sogues & Luis Serrano. A reporter system coupled with high-throughput sequencing unveils key bacterial transcription and translation determinants. Nature Communications, 8, 368 (2017). DOI: 10.1038/s41467-017-00239-7.
Funding information:
This work was supported by the Fundación Marcelino Botín and the Spanish Ministerio de Economía, Industria y Competitividad (BIO2007-61762), the Instituto de Salud Carlos III, the Federación Española de Enfermedades Raras (PI10/01702), the European Research Council (ERC) under the European Union’s Horizon 2020 program (634942 – MycoSynVac and 670216 – Mycochassis), the Centro de Excelencia Severo Ochoa 2013-2017 grant, the Juan de la Cierva Program (IJCI-2014-22070).
Media Contact:
Laia Cendrós, Media Relations, Centre for Genomic Regulation (CRG) - Tel.+34 93 316 02 37
| Adjunto | Tamaño |
|---|---|
| 103.2 KB | |
| 105.11 KB | |
| 107.29 KB |

