NOTÍCIES
NOTÍCIES
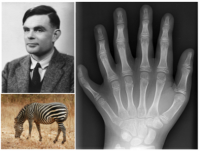
CRG researchers confirm that a mathematical theory first proposed by Alan Turing in 1952 can explain the formation of fingers
PRESS RELEASE
- In a paper published today in Science, researchers from the Centre for Genomic Regulation (CRG) in Barcelona, show that BMP and WNT proteins are the so-called “Turing molecules” for creating embryonic fingers
- This confirms a fundamental theory first proposed by the founding father of computer science, Alan Turing, back in 1952.
- BMPs and WNTs interact in a self-organising process, producing a repetitive pattern of gene expression that determines which cells should become fingers.
- This explains why polydactyly – the development of extra fingers or toes – is relatively common in humans, affecting up to 1 in 500 births
Barcelona, 31st of July 2014. Alan Turing, the British mathematician (1912-1954), is famous for a number of breakthroughs, which altered the course of the 20th century. In 1936 he published a paper, which laid the foundation of computer science, providing the first formal concept of a computer algorithm. He next played a pivotal role in the Second World War, designing the machines which cracked the German military codes, enabling the Allies to defeat the Nazis in several crucial battles. And in the late 1940’s he turned his attention to artificial intelligence and proposed a challenge, now called the Turing test, which is still important to the field today.
His contribution to mathematical biology is less famous, but was no less profound. He published just one paper (1952), but it triggered a whole new field of mathematical enquiry into pattern formation. He discovered that a system with just 2 molecules could, at least in theory, create spotty or stripy patterns if they diffused and chemically interacted in just the right way.
His mathematical equations showed that starting from uniform condition (ie. a homogeneous distribution – no pattern) they could spontaneously self-organise their concentrations into a repetitive spatial pattern. This theory has come to be accepted as an explanation of fairly simple patterns such as zebra stripes and even the ridges on sand dunes, but in embryology it has been resisted for decades as an explanation of how structures such as fingers are formed.

Now a group of researchers from the Multicellular Systems Biology lab at the CRG, led by ICREA Research Professor James Sharpe, has provided the long sought-for data which confirms that the fingers and toes are patterned by a Turing mechanism. "It complements our recent paper (Science 338:1476, 2012), which provided evidence that Hox genes and FGF signaling modulated a hypothetical Turing system. However, at that point the Turing molecules themselves were still not identified, and so this remained as the critical unsolved piece of the puzzle. The new study completes the picture, by revealing which signaling molecules act as the Turing system" says James Sharpe, co-author of the study.
The approach taken was that of systems biology – combining experimental work with computational modelling. In this way, the two equal-first authors of the paper were able to iterate between the empirical and the theoretical: the lab-work of Jelena Raspopovic providing experimental data for the model, and the computer simulations of Luciano Marcon making predictions to be tested back in the lab.
By screening for the expression of many different genes, they found that two signalling pathways stood out as having the required activity patterns: BMPs and WNTs. They gradually constructed the minimal possible mathematical model compatible with all the data, and found that the two signalling pathways were linked through a non-diffusible molecule – the transcription factor Sox9. Finally, they were able to make computational predictions about the effects of inhibiting these 2 pathways – either individually, or in combination – which predicted how the pattern of fingers should change. Strikingly, when the same experiments were done on small pieces of limb bud tissue cultured in a petri dish the same alterations in embryonic finger pattern were observed, confirming the computational prediction.
"Arriving at the correct understanding of multicellular organization is essential if we are to develop effective strategies for regenerative medicine, and one day to possibly engineer replacement tissues for various organs"
This result answers a long-standing question in the field, but it has consequences that go beyond the development of fingers. It addresses a more general debate about how the millions of cells in our bodies are able to dynamically arrange themselves into the correct 3D structures, for example in our kidneys, hearts and other organs. It challenges the dominance of an important traditional idea called positional information, proposed by Lewis Wolpert which states that cells know what to do because they all receive information about their “coordinates” in space (a bit like longitude and latitude on a world map). Today’s publication highlights instead that local self-organising mechanisms may be much more important in organogenesis than previously thought.
Arriving at the correct understanding of multicellular organization is essential if we are to develop effective strategies for regenerative medicine, and one day to possibly engineer replacement tissues for various organs. In the shorter term, these results also explain why polydactyly – the development of extra fingers or toes – is such a common birth defect in humans: Turing systems are mathematically known to have slightly lower precision in regulating the number of “stripes” than alternative models.
At first glance, the question of how an embryo develops seems unrelated to the problems of computing and algorithms with which Turing is more commonly associated. In reality however, they were both expressions of his interest in how complex and clever biological “machines” arise in nature. In a sense, he sought the algorithms by which life builds itself. It is fitting that this study, which has confirmed Turing’s 62 year-old theory on embryology, required the development of a serious computer model. It brings together two of his major life achievements into one satisfying result.
For further information: Juan Manuel Sarasua, Press Office at the CRG - juan.sarasua@crg.eu
Related Article:
J. Raspopovic; L. Marcon; L. Russo; J. Sharpe. Digit patterning is controlled by a Bmp-Sox9-Wnt Turing network modulated by morphogen gradients. Science, 2014. DOI: 10.1126/science.1252960
Related links:
-How Did You Get Five Fingers? by Ed Yong, in "Not Exactly Rocket Science" (National Geographic): http://phenomena.nationalgeographic.com/2014/08/01/how-did-you-get-five-fingers/
Biologists Home in on Turing Patterns. Was Alan Turing right about the mechanism behind tiger stripes? (Quanta Magazine, http://www.simonsfoundation.org/quanta/20130325-biologists-home-in-on-turing-patterns/)
"Turing patterns show their hand in finger formation". Article by Andy Stance in Chemistry World: http://www.rsc.org/chemistryworld/2014/07/turing-patterns-show-hand-finger-formation
"Alan Turing también descifró el código oculto de cómo se forma el cuerpo". Por Nuño Dominguez en Materia: http://esmateria.com/2014/07/31/alan-turing-tambien-descifro-el-codigo-oculto-de-como-se-forma-el-cuerpo-humano/
TEDxBarcelona - James Sharpe - Digital digits: Revealing Nature's cellular computers (subtl spanish: http://youtu.be/CtiZeTPzExE)
A. M. Turing. The Chemical Basis for Morphogenesis . Philosophical Transactions of the Royal Society of London. Series B, Biological Sciences, Vol. 237, No. 641. (Aug. 14, 1952), pp. 37-72.
Cover photo credits:
"Alan Turing photo" by http://www.ieee.org/portal/cms_docs_sscs/sscs/08Spring/KFig6_turing.jpg. Licensed under Fair use of copyrighted material in the context of Alan Turing via Wikipedia - http://en.wikipedia.org/wiki/File:Alan_Turing_photo.jpg#mediaviewer/File...
"Equus quagga boehmi (grazing)" by Hans Hillewaert - Own work. Licensed under Creative Commons Attribution-Share Alike 3.0 via Wikimedia Commons - http://commons.wikimedia.org/wiki/File:Equus_quagga_boehmi_(grazing).jpg#mediaviewer/File:Equus_quagga_boehmi_(grazing).jpg
"Polydactyly 01 Lhand AP" by en:User:Drgnu23, subsequently altered by en:user:Grendelkhan, en:user: Raul654, and en:user:Solipsist. - Own work. Licensed under Creative Commons Attribution-Share Alike 3.0 via Wikimedia Commons - http://commons.wikimedia.org/wiki/File:Polydactyly_01_Lhand_AP.jpg#media...
| Attachment | Size |
|---|---|
| 108.24 KB |