NEWS
NEWS
Humans and Mice: Similar enough for studying disease and different enough to give us new clues about evolution
- An international consortium including researchers from the Centre for Genome Regulation led by Roderic Guigó has presented an exhaustive description of the mouse's functional genome elements and their comparison with the human genome.
- The work at the CRG was carried out in close collaboration with the group directed by Dr. Thomas R. Gingeras, at the Cold Spring Harbour Laboratory in the United States.
- Comparing humans and mice enables us to better understand mammalian biology and evolution, as well as contributing new information on the use of mice as animal models for looking at human disease.
- The results will be published on November 20 in four articles in the journal Nature as well as others in various different scientific publications.
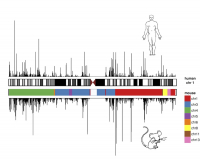
A group of international researchers has just discovered the keys to explaining why certain processes and systems in mice, like the immune system, metabolism and stress response, are so different to those in humans. The scientists have detailed the functional parts of the mouse genome and have compared them with those in humans. A whole set of data has come out of this – which is now to available to the scientific community – which will be significant for research into mammalian biology as well as the study of human illness mechanisms.
The comparison focuses on the genetic and biochemical processes regulating genome activity in humans and mice. The scientists have found that, in general, the systems for controlling genome activity in the two species are very alike, and have been preserved through time. However, they have also detected certain differences in the DNA, and patterns of gene expression that are not shared. “Finding these similarities and studying the aspects of mouse biology that may reflect human biology, allows us to approach the study of human illnesses in a better way”, affirms Bing Ren, one of the principal authors from the ENCODE Consortium and a lecturer in molecular and cellular medicine at the University of California – San Diego.
“The mouse is one of the most utilised models for studying human biology and we use it for creating models of human illnesses and testing new drugs and therapies. Our study goes a long way towards validating the usefulness of this animal model and provides enormous support for its use in human illnesses. We have found that there are many well-preserved cell processes in the two species, for example, in embryonic development. Understanding these similarities will allow us to carry out more accurate studies on human biology”, explains Roderic Guigó, one of the main researchers involved in the work and coordinator of the Bioinformatics and Genomics programme at the CRG.
The researchers have compared various processes involved in gene expression, such as gene transcription and chromatin modification, and have repeated this in different tissues and cell types from both humans and mice. “Our lab took part in analysing the group of RNA or transcriptome, that results from transcription, the process by which the instructions in the genes are read. We have discovered that human and mice transcriptome contains both preserved and divergent elements. Surprisingly we have found that the differences seem bigger between species rather than between tissues when initially we thought that the gene activity in the same kinds of tissues would be similar”, adds Alessandra Breschi, one of the first co-authors of the main work published in Nature and a researcher in Roderic Guigó’s lab at the CRG.
The project makes it clear that there is a wide variety of options available for achieving gene expression. By comparing these two genomes they have also found that there is a common “language” that the cells use at the molecular level but which, at the same time, is tremendously flexible and has varied greatly throughout evolution. For example, if we used the analogy of electrical circuits, we would find cables, plugs, switches, etc. By combining the pieces in one way or another, we would obtain very different circuits (as happens between mice and humans) although the basic mechanisms governing the operation are based on the same methods and available resources.

An additional study, currently available at bioRxiv, led by the researchers from the CRG and Cold Spring Harbour Laboratory, highlights the fact that a substantial part of human and mice genes have maintained an essentially constant expression throughout evolution, in tissues and various organs. In addition, the researchers have quantified the preservation level of this gene expression between humans and mice. This allows the identification of those genes that have the same expression in the two species, and for which the mouse represents a good model of human biology.
“ENCODE is a living project and the maps that are generated are constantly updated and improved, with information being added on new types of cells and tissues or new complementary genome assays. We hope that the project can keep providing this data as it has up to now, making it available to everyone and treating it in a systematic and coherent way”, concludes Dr. Guigó, the only principal investigator in Europe involved in this work.
The results of the comparison will be published on November 19 in four articles in the prestigious journal Nature. As well as this, other scientific journals including Genome Research, Genome Biology, PNAS, Blood and Nature Communications are also bringing out related articles. The ENCODE Consortium, or ENCyclopedia of DNA Elements, is a research programme on the human genome funded by the US National Institutes of Health.
References:
Articles published in the journal Nature on November 20, 2014:
- A comparative encyclopedia of DNA elements in the mouse genome DOI: 10.1038/nature13992
- Conservation of trans-acting circuitry during mammalian regulatory evolution DOI: 10.1038/nature13972
- Principles of regulatory information conservation between mouse and human DOI: 10.1038/nature13985
- Topologically associating domains are stable units of replication-timing regulation DOI: 10.1038/nature13986
For further information and interviews:
Centre for Genomic Regulation – Press Office
Laia Cendrós - Tel. 93 316 0237
| Attachment | Size |
|---|---|
| 128.34 KB | |
| 139.41 KB |